Use the
Spin on button at the bottom of the model window to rotate the models at any time.
Alternatively, you can rotate either using the mouse, or by right clicking on the image, and selecting
Spin > On from the menu.
Stop spinning by right clicking on the image and selecting
Spin > Off from the menu.
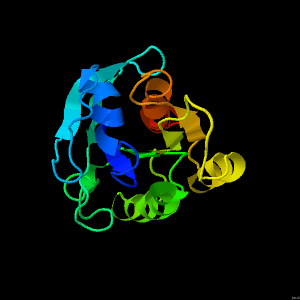
Ras in involved in the control of cellular proliferation and differentiation. Mutations of the ras protein are involved in 30% of human cancers.
Thus, it is very important to understand the structure and function of this enzyme.
The ras molecule is made up of 5 alpha helices (in Pink) and a six-stranded beta sheet (in Green).
Spin the model to observe the arrangement. In the inactive state, Ras is bound to GDP.
Spin the model to observe the arrangement. In the activated ras, GTP is bound. It is held between two flexible chains known as Switch I (in Aqua) and Switch II (in Magenta).
Spin the model to observe the arrangement.These switches undergo conformational changes upon the transition from the GTP-bound state to the GDP-bound state.
Switch I has 5 acidic residues (in Yellow - 3 aspartic acid and 2 glutamic acid residues) that create a negatively charged surface which is used to interact with effector proteins (proteins that respond to activation of ras and propagate the signal).
Spin the model to observe the arrangement.Ras is inactivated when the bound GTP is hydrolyzed to GDP. This reaction occurs very slowly in ras and is stimulated by GTPase activating proteins, or GAPs. GAP inserts its arginine finger (Arg789) into the active site, stabilizing the developing charge on the gamma phosphate of GTP thereby stabilizing the transition state and catalysing the hydrolysis of GTP to GDP.
The ras Switches hold the attacking water molecule and gamma phosphate in exactly the right position for hydrolysis to occur.Spin the model to observe the arrangement.
Switch II allows Gln61 (in Purple) to participate in catalysis.
Spin the model to observe the arrangement.
Gln61 is hydrogen bonded to the arginine finger and functions to stabilize the GTPase reaction transition state. Mutations in Gln61 block hydrolysis and lead to tumor formation.
Spin the model to observe the arrangement.
Gly12 (in Green) is in van der Waals contact with the arginine finger. Any residue other than glycine would displace the arginine finger— preventing its interaction with the gamma phosphate thereby rendering GAP unable to hydrolyze GTP and unable to turn off the ras-mediated mitogenic signal. Thus, mutations in Gly12 also oncogenic.
Currently, researchers are trying to design drugs that will act as a surrogate finger which will insert into the active site, stimulate GTP hydrolysis, and turn off the ras signal.
Spin the model to observe the arrangement.
You may look at any of these intermediate views again by clicking on the appropriate button.
Based on a template by A. Herráez and J. Gutow
Using directory /Users/George/Projects/JMol/RAS - Out/ras
adding support.js
...jmolApplet0
...adding Reset.png
copying and unzipping jsmol.zip directory into /Users/George/Projects/JMol/RAS - Out/ras
...copying
/Users/George/Projects/JMol/RAS/ras-fixed.pdb.gz
to
/Users/George/Projects/JMol/RAS - Out/ras/ras-fixed.pdb.gz
...adding Reset.spt
...jmolApplet1
...adding Rotate.png
copying and unzipping jsmol.zip directory into /Users/George/Projects/JMol/RAS - Out/ras
...adding Rotate.spt
...jmolApplet2
...adding Colour_secondary_structure.png
copying and unzipping jsmol.zip directory into /Users/George/Projects/JMol/RAS - Out/ras
...adding Colour_secondary_structure.spt
...jmolApplet3
...adding Show_bound_GTP.png
copying and unzipping jsmol.zip directory into /Users/George/Projects/JMol/RAS - Out/ras
...adding Show_bound_GTP.spt
...jmolApplet4
...adding Show_Switch_regions.png
copying and unzipping jsmol.zip directory into /Users/George/Projects/JMol/RAS - Out/ras
...adding Show_Switch_regions.spt
...jmolApplet5
...adding Show_acidic_residues_on_Switch_I.png
copying and unzipping jsmol.zip directory into /Users/George/Projects/JMol/RAS - Out/ras
...adding Show_acidic_residues_on_Switch_I.spt
...jmolApplet6
...adding Flicker_switches.png
copying and unzipping jsmol.zip directory into /Users/George/Projects/JMol/RAS - Out/ras
...adding Flicker_switches.spt
...jmolApplet7
...adding Show_Gln61.png
copying and unzipping jsmol.zip directory into /Users/George/Projects/JMol/RAS - Out/ras
...adding Show_Gln61.spt
...jmolApplet8
...adding Show_Gly12.png
copying and unzipping jsmol.zip directory into /Users/George/Projects/JMol/RAS - Out/ras
...adding Show_Gly12.spt